.png?width=815&height=379&name=Frame%20160%20(1).png)
Proteomics International offers biomarker discovery, verification, and analytical validation services through its Promarker® technology platform—a powerful mass spectrometry-based approach for identifying protein biomarkers.
Our ISO 17025-accredited laboratory provides these services as a fee-for-service model or through collaborative partnerships, supporting pharmaceutical, biotech, and research organizations in accelerating biomarker-driven innovations.
The Promarker® platform has successfully identified and validated protein biomarkers, including those for diabetic kidney disease, which led to the development of the Promarker®D test. This expertise is now available to clients looking to enhance their drug development, clinical trial stratification, and precision medicine initiatives.
Partner with us to leverage our advanced mass spectrometry capabilities and expertise in biomarker discovery/ validation.
Biomarker Discovery
(Labelled Quantitation – Tandem Mass Tag [TMT] Technology)
Proteomics International offers advanced biomarker discovery and proteome quantitation services using Tandem Mass Tag (TMT) technology.
TMT for Quantitative Proteomics
TMT-based proteomics enables:
- Simultaneous quantitation of up to multiple samples in a single mass spectrometry experiment
- Accurate relative quantitation of proteins and peptides
- Detection of differential protein expression across experimental conditions
- Enhanced sensitivity for low-abundance proteins
- Streamlined workflows for clinical research, biomarker validation, and drug development
Sample Requirements
To ensure optimal results:
- Protein Input: 200–500 µg recommended; minimum 50 µg
- Sample Types: Cell lysates or secreted proteins (serum/plasma-free media)
- Volume: Samples should be <250 µL
- Protease Inhibitors: Recommended
- Genome Availability: Required for accurate protein identification and quantitation
Our team of experts work closely with clients to ensure tailored experimental design, high-quality mass spectrometry data, and insightful bioinformatics analysis.
Biomarker Validation
(Targeted Mass Spectrometry – MRM/SRM Quantitation)
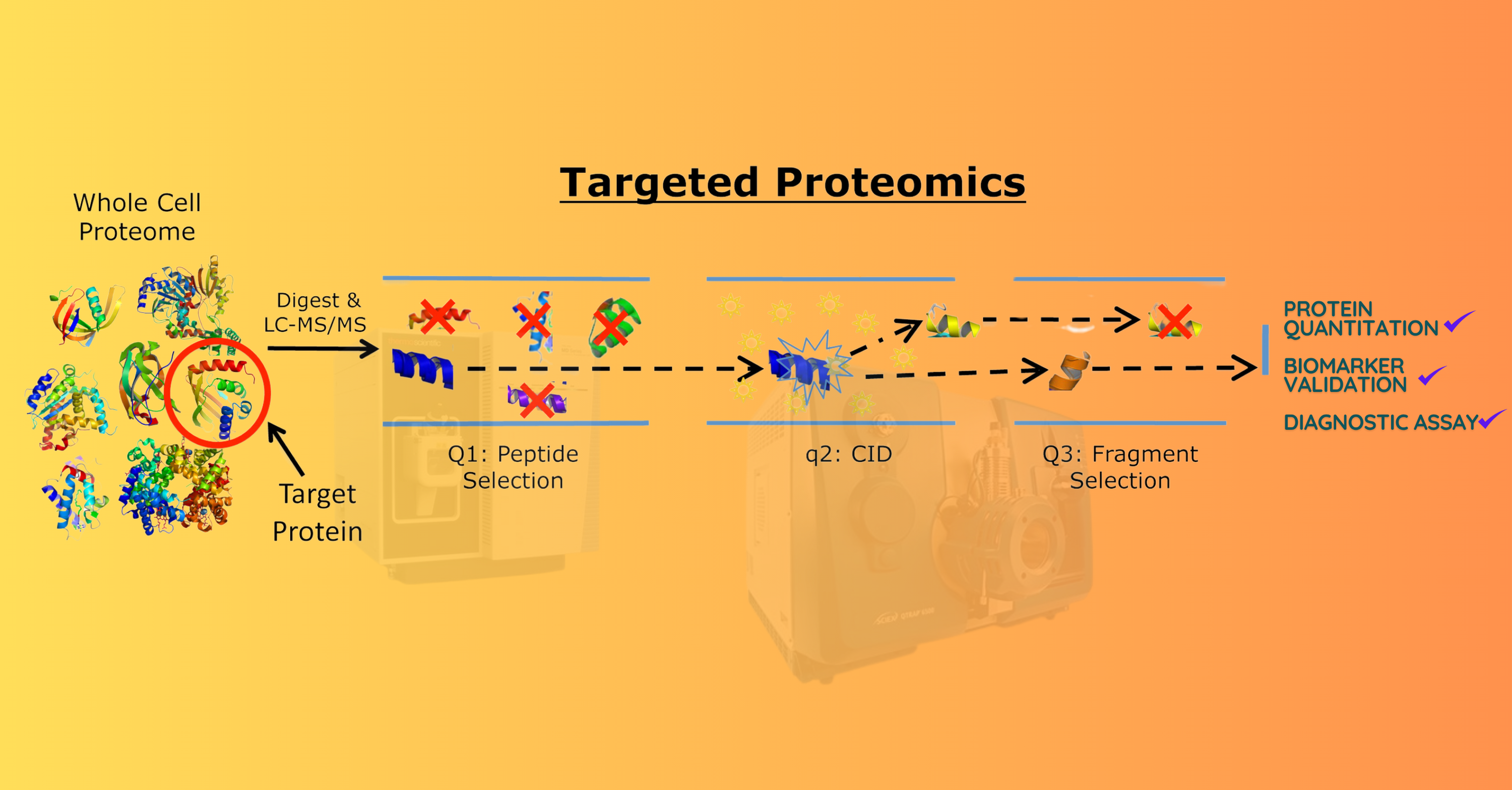
Proteomics International offers expert biomarker validation services using targeted mass spectrometry.
MRM for Targeted Quantitation
Targeted proteomics using MRM/SRM allows:
- Highly specific and sensitive quantitation of target peptides
- Reproducible measurement of protein biomarkers across sample cohorts
- Development of custom assays for protein signature validation
- Efficient method transfer to clinical and diagnostic settings
- Quantitative analysis compatible with serum, plasma, cell lysates, and other biological matrices
Applications
- Verification of protein biomarkers from TMT-based discovery
- Pharmacodynamic monitoring in preclinical and clinical studies
- Support for drug mechanism-of-action, patient stratification, and disease progression analysis
- Development of targeted assays for clinical validation or diagnostic use
Proteomics International’s ISO 17025-accredited laboratory ensures analytical robustness and regulatory compliance.
Resources
Technical Technote:
- Protein Biomarker Research Pipeline for Developing Protein Biomarkers for Diabetic Kidney Disease
- Targeted Mass Spectrometry for Biomarker Validation – MRM/SRM Overview (PDF)
Relevant Publications
Table of Contents
Application Examples
Protein Quantitation MRM
Reference for article on MRM of peptides to determine relative protein quantitation: Expression of Cardiac a-Actin Spares Extraocular Muscles in Skeletal Muscle a-Actin Diseases
This article describes the development and implementation of an MRM based method to determine relative quantities of two proteins (cardiac and skeletal actin) which differ in only 4 residues out of 375 in their sequences, i.e. they are 99% homolgous. The proteins were processed together and specific MRMs developed that would detect the presence of each form sensitively and specifically. Only relative quantitation was required for this application.
Peptide/Lipopeptide Quantitation Using MRM
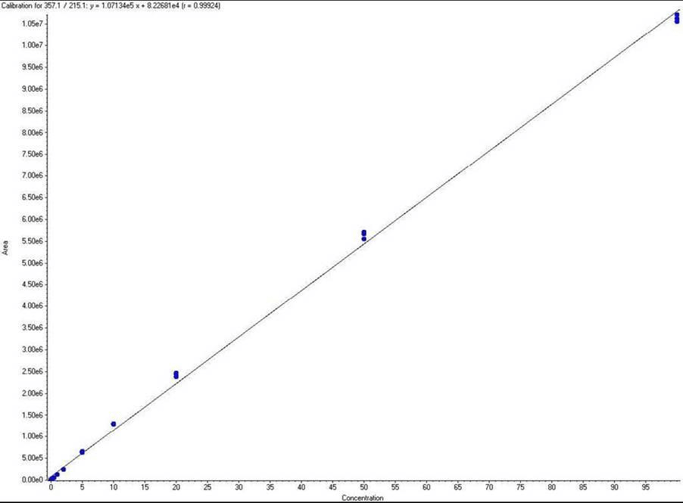
FAQ
What types of samples can be analysed using MRM?
Multiple Reaction Monitoring (MRM) analysis is highly versatile and suitable for a wide range of sample types. Any compound that can be ionised and fragmented is a candidate for MRM quantitation. The most common applications include the analysis of proteins, peptides, metabolites, and small molecule drugs in complex biological matrices such as plasma, serum, or tissue extracts.
MRM is especially effective for targeted protein quantitation using digested peptides that represent the protein of interest. This approach is widely used in biomarker validation, as it enables accurate and reproducible detection in complex clinical samples. Multiple transitions can be monitored in a single run, allowing simultaneous quantitation of multiple analytes.
How sensitive is MRM?
MRM is a highly sensitive mass spectrometry technique capable of detecting compounds at extremely low concentrations. For example, peptides can be quantitated in the femtomole range, and in complex biological fluids such as plasma, quantitation at microgram per millilitre (µg/mL) levels is achievable. This high sensitivity makes MRM an ideal method for detecting low-abundance biomarkers and monitoring subtle changes in protein expression.
Can MRM be used for absolute and relative quantitation?
Yes. MRM supports both absolute and relative quantitation of proteins and peptides.
- Relative quantitation compares changes in protein abundance across samples.
- Absolute quantitation requires spiking samples with known quantities of synthetic stable isotope-labelled peptide standards, which serve as internal references to calculate exact protein concentrations.
Is method development required for MRM analysis?
Yes. Each MRM assay is custom-developed, requiring careful optimisation of instrument parameters and peptide transitions. This includes selecting optimal parent-to-fragment ion pairs, calibrating sensitivity, and, if necessary, applying sample cleanup or chromatographic separation.
For complex matrices like plasma or serum, additional sample preparation steps are required, which may impact turnaround time and cost.
Download Biomarker Discovery & Validation Flyer
